Research
Research in the Teitell group focuses on mechanisms of cancer formation and progression. We place a special emphasis on studies of leukemia and lymphoma that arise during normal B cell development. Our work has evolved into several areas with distinct sets of techniques and approaches; however, all project areas address fundamental questions concerning the etiology and progression of cancer, providing a common link for the group. Areas of active research in our group include:
B Lymphocyte Development and Cancer
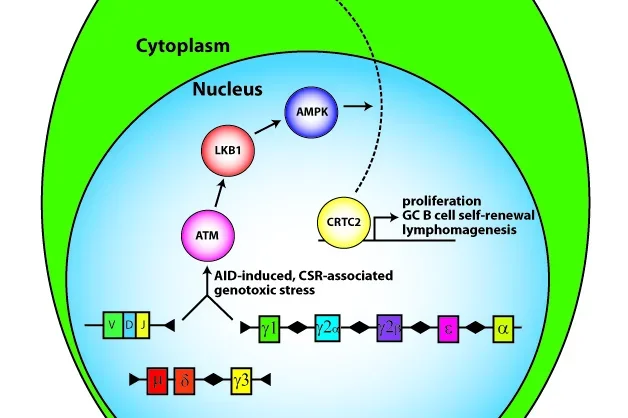
Our group discovered a role for aberrant TCL1 oncogene expression in human leukemias and lymphomas and generated a proof-of-principle TCL1 transgenic mouse model that develops mainly germinal center (GC) B cell cancers (Teitell PNAS 1999; Hoyer PNAS 2002; Teitell Nature Reviews Cancer 2005). Studies of TCL1 gene regulation in B cell development and cancer ( PNAS 2007) led to the identification of a signal transduction pathway that controls the terminal differentiation of GC B cells into antibody-secreting plasma cells (Sherman Molecular Cell 2010; Sherman Trends in Cell Biology 2011). Key steps in this signal transduction pathway include the activation of the tumor suppressor LKB1 and that of an unknown AMPK (5’ AMP activated protein kinase) family member protein, which results in the nuclear export and inactivation of the CREB transcriptional co-activator, CRTC2 (figure). Inactivation of CRTC2 represses direct target genes to enable B cells to exit the GC reaction and differentiate into plasma cells. Failure to inactivate CRTC2 blocks differentiation and locks down GC B cells in a state of continued growth and increased resistance to apoptosis, thereby generating an environment that fosters tumor formation. Surprisingly, this signal transduction pathway also suppressed the premature formation of GCs by inhibiting spontaneous B cell activation (Walsh EMBO Reports 2015). In sum, we uncovered and continue to study a signal transduction pathway and genetic program that controls the initiation, progression, and termination of an effective humoral immune response (Waters Cell Cycle 2015). Errors in this signal transduction pathway or genetic program provide known and novel candidates for uncontrolled B cell growth, survival, and malignant transformation.
Current lab project areas include:
(Techniques include ChIP, next-generation sequencing, epigenetic approaches, IP-mass spectrometry, mouse models, quantitative imaging, and many standard cellular and molecular biology approaches)
(1) Dissection of CRTC2 regulation of B lymphocyte activation, differentiation, and function
(2) Evaluation of a mouse model that is defective for antigen-driven plasma cell differentiation
(3) Molecular elucidation of the role for LKB1 and AMPK in starting GC reactions
Linking RNA Processing with Mitochondrial Homeostasis, Metabolism, and Proliferation
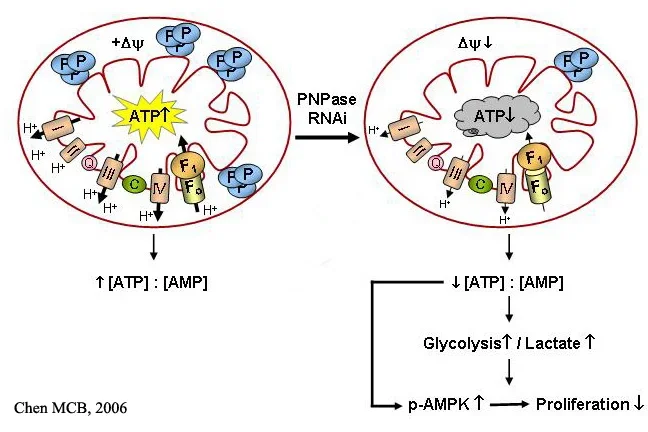
A mass spectrometry search for proteins that interact with the TCL1 oncoprotein to potentially promote cancer yielded polynucleotide phosphorylase (PNPase) as a promising candidate ( Cancer Lett 2006) . PNPase is an evolutionarily conserved exoribonuclease that unexpectedly localizes in the intermembrane space of mammalian mitochondria ( MCB 2006; MCB 2006) . We determined that PNPase supports respiration, maintains mitochondria homeostasis with an unknown role in affecting fusion versus fission, regulates energy metabolism, and controls cell proliferation ( MCB 2006; MCB 2006) . The interaction with TCL1, which is an AKT coactivator and stimulates signaling in the AKT and PKC/MAPK/ERK pathways ( Biochemistry 2002; J Immunol 2005) , potentially links control of mitochondria function to major signal transduction pathways implicated in a large variety of human malignancies. Current studies include determining the molecular mechanisms for the effect of PNPase on mitochondria structure and function and the role that altered metabolism plays in supporting malignancy within the immune system.
Nanoscale Evaluation of Malignant Transformation
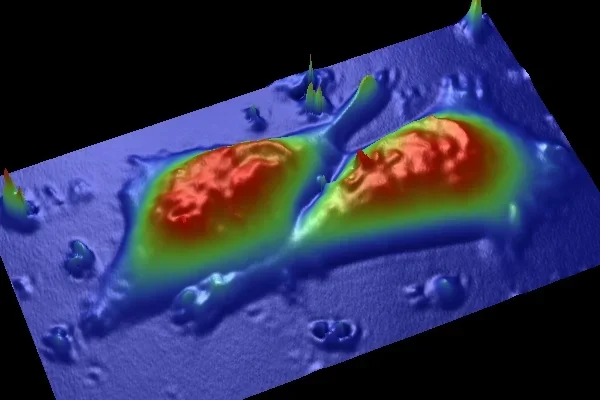
Our group has been examining the nanoscale structural and functional characteristics of cells with an atomic force microscope (AFM) ( Nanomedicine 2006) and micron sized particles embedded within cells using bio-microrheology techniques ( Biophys J 2006; Microfluidics & Nanofluidics 2007). AFM and particle tracking methods enable detection of the changes in cells during their progression toward malignancy or increasingly aggressive malignant behavior. Our methods help determine cell damage before biochemical evidence of damage is obtained, with current studies targeting the processes that regulate structural changes at the membrane and within the cytoplasm, to provide a novel, integrated view of cancerous transformation.
To further support these studies, we are co-recipients of a Nanomedicine Development Center (NDC) Award from the NIH Nanomedicine Roadmap Initiative. Ongoing studies include:
- a collaboration to further the development of optoelectronic tweezers (OET) technology for moving cells and subcellular structures without damage for multiplex single cell analysis;
- a collaboration to develop interferometric microscopy with cell nanomirrors to evaluate malignant progression and identify “outlier” cells in a pool of seemingly identical cancer cells that may be particularly harmful ( Nanotechnology 2007);
- a collaboration to identify specific DNA molecules in complex mixtures using AFM techniques (Reed Nanotechnology 2007);
- an NDC collaboration to develop quantitative technologies for evaluating signal transduction kinetics and strength in live cells; and
- a collaboration to develop a new type of live cell surgery to facilitate the repair and manipulation of somatic and approved human embryonic stem cells and their derivatives.
Mitochondria in Human Embryonic Stem Cell Self-Renewal vs. Differentiation
A new area of investigation links developing expertise in the group in two areas- mitochondrial and human embryonic stem cell (hESC) biology. The rationale for these studies is the need to understand how changes in metabolism, which normally shifts from glycolysis to aerobic respiration as fertilized ova implant during embryogenesis, shifts back again to a main dependence on glycolysis (the Warburg effect) with malignant transformation. We reason that dynamic changes in mitochondria function and hESC dependence on oxygen tension will mimic key features of malignant degeneration. Current studies include evaluating the role of native and damaged mitochondria in normoxic and hypoxic oxygen tension environments to determine a role in controlling hESC decisions for self-renewal, differentiation, or quiescence, which could resemble similar decisions in “cancer stem cells”.
Human Embryonic Stem Cells and Cancer
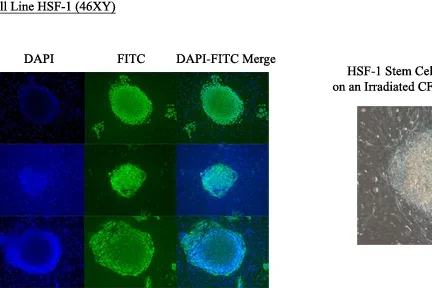
Mouse Tcl1 is the only known member of an oncogene family thus far shown to control basic embryonic stem cell decisions, such as self-renewal. Human TCL1 family members are expressed in early embryonic development, in skin appendage stem cells, and in germ cell tumors, which share many characteristics with hESCs. Furthermore, the major signaling pathway that controls hESC survival is the PI3K/AKT pathway, which TCL1 coactivates. Since aberrant TCL1 expression causes models for major human leukemias and lymphomas in transgenic mice ( PNAS 2002; Nature Rev Cancer 2005; Blood 2006) , and many human germ cell tumors and germinal center derived Burkitt lymphomas demonstrate aberrant TCL1 expression ( AJCP 2005) , we reason that TCL1 family member expression in hESCs could controls decisions regulating self-renewal, differentiation, survival, and quiescence. Understanding this regulation should provide clues to the malignancy-inducing mechanism of aberrant TCL1 expression in human cancers and perhaps insight into the pathophysiology of “cancer stem cells”. Current studies include manipulations of TCL1 family expression levels in hESCs with determination of effects decisions to proliferate, apoptose, or differentiate.